Setup Cellpose integration
Table Of Contents

1.74.x+
This guide was written for JIPipe version 1.74.0 or newer
Quick setup
JIPipe features an integration of Cellpose version 2.x. This software is not part of ImageJ and its update site ecosystem and thus requires additional installation steps. These steps are generally only necessary if JIPipe notifies you about the missing installation of the third-party software.
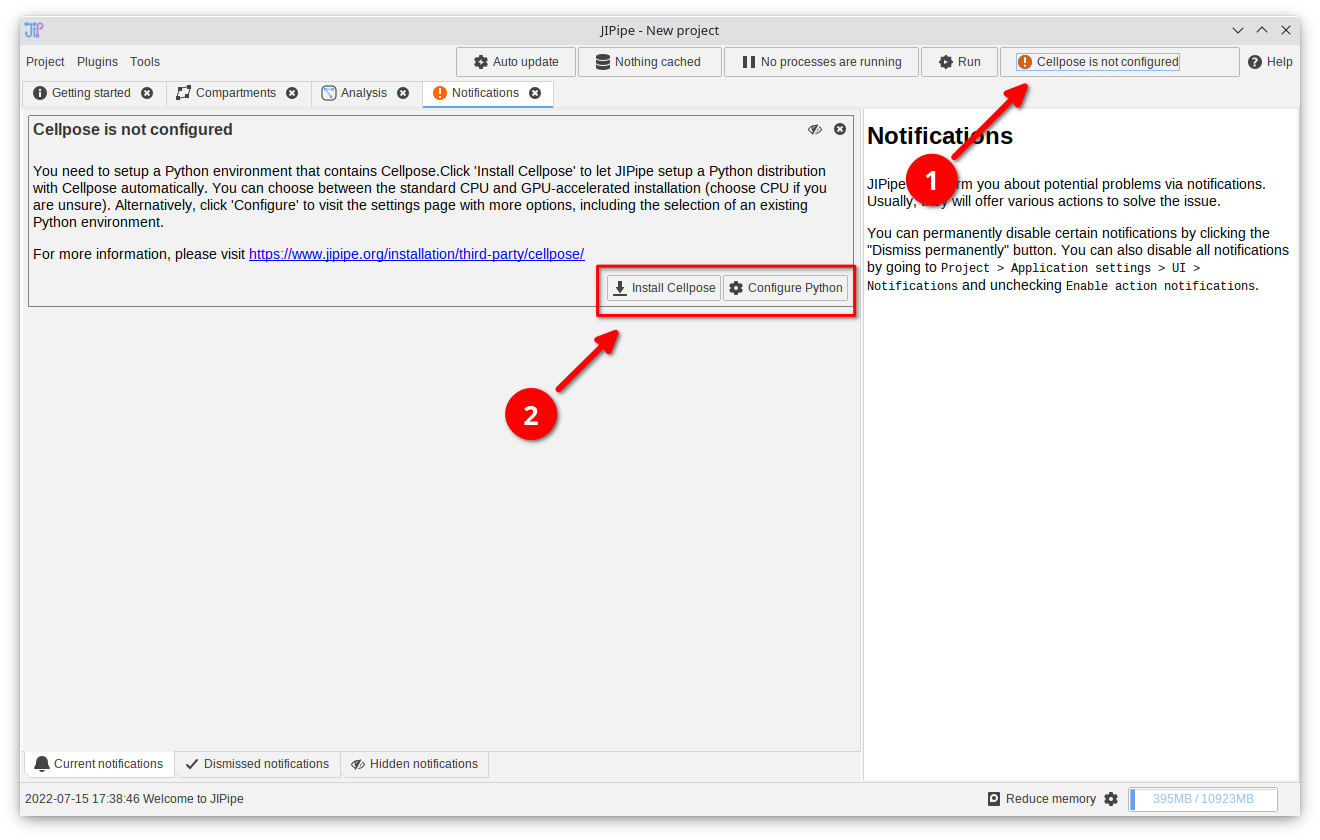
Notification: Cellpose not configured
If JIPipe was not yet configured with a Cellpose Python environment, you will be notified on opening the application. Click the notifications button and select the "Install Cellpose" option and follow the instructions or choose "Configure Python".
You can always change the Python environment by navigating to
Project > Application settings > Extensions > Cellpose if you clicked away the notification or if you want to make further changes.
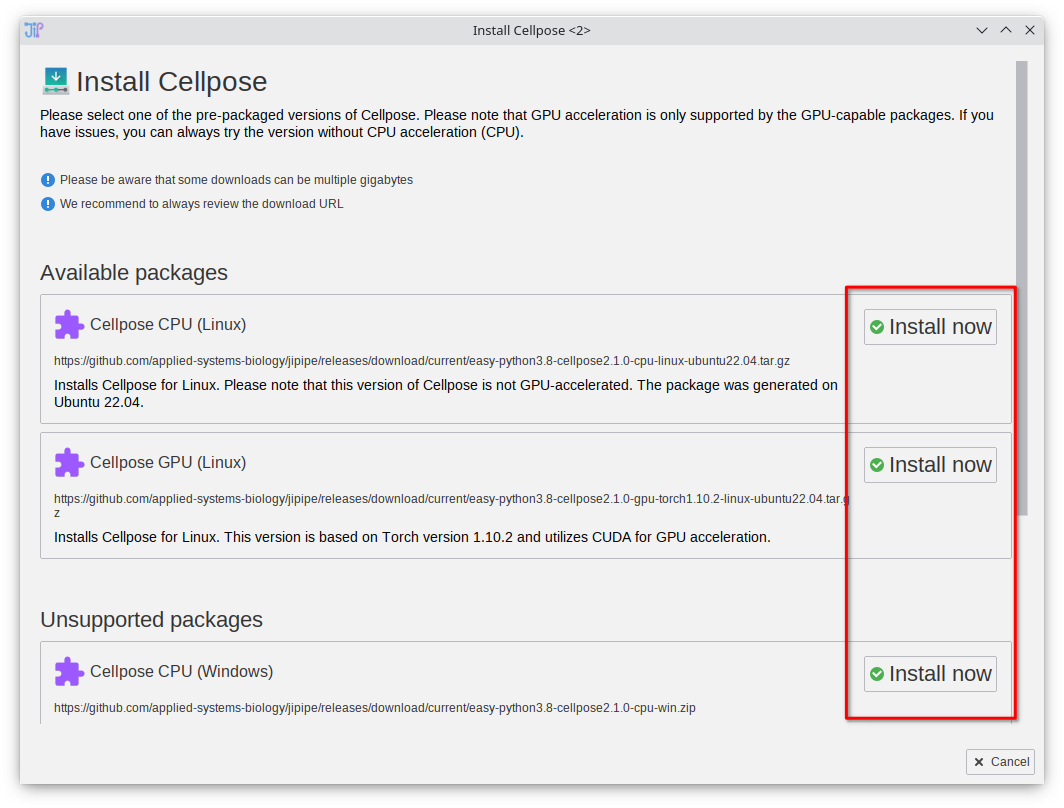
Install Cellpose (prepackaged)
On selecting the "Install Cellpose" option, JIPipe will provide you with a pre-made environment that is installed into the ImageJ directory. Choose any of the supported packages. If you already have existing Python environments with Cellpose installed, please follow the other tutorials on this page.
MacOS: We were not yet successful in generating a pre-packaged GPU-accelerated version of Cellpose for MacOS.
GPU accelerated versions: We created the packages with Pytorch for CUDA 10.2. Your system might require a different setup (see pytorch documentation). In this case, please install Cellpose via our automated installer or follow the official Cellpose installation tutorial to create a Conda environment that can be selected within JIPipe.
Selecting an existing Conda environment
Cellpose utilizes JIPipe's Python environment system and is compatible to the existing installation options provided by the Python integration.
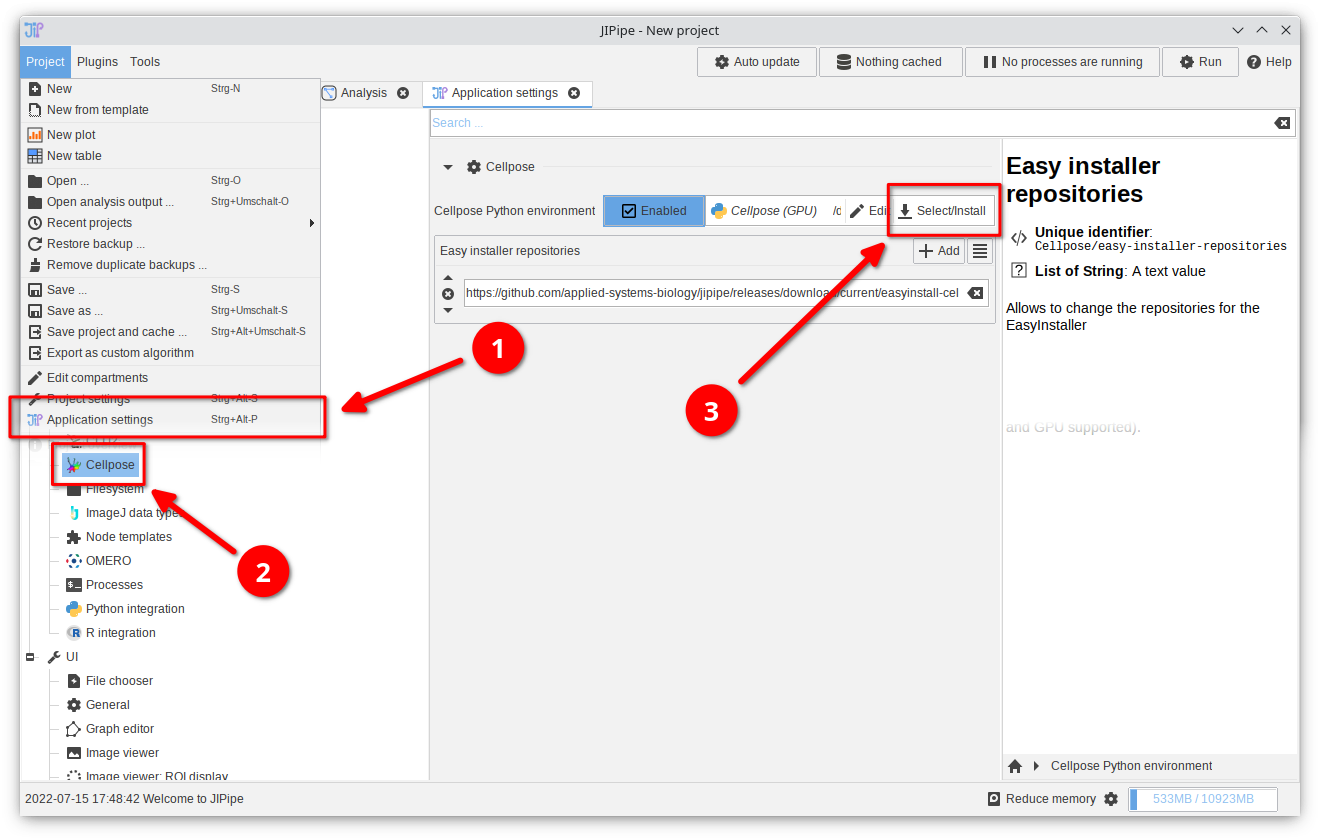
1. Open the Python integration settings
Unless you clicked "Configure Python" in the notification panel, navigate toProject > Application settings > Extensions > Cellpose.
Click the "Select/Install" button in the "Python environment" setting and choose Additional compatible installers > Select existing Conda environment.
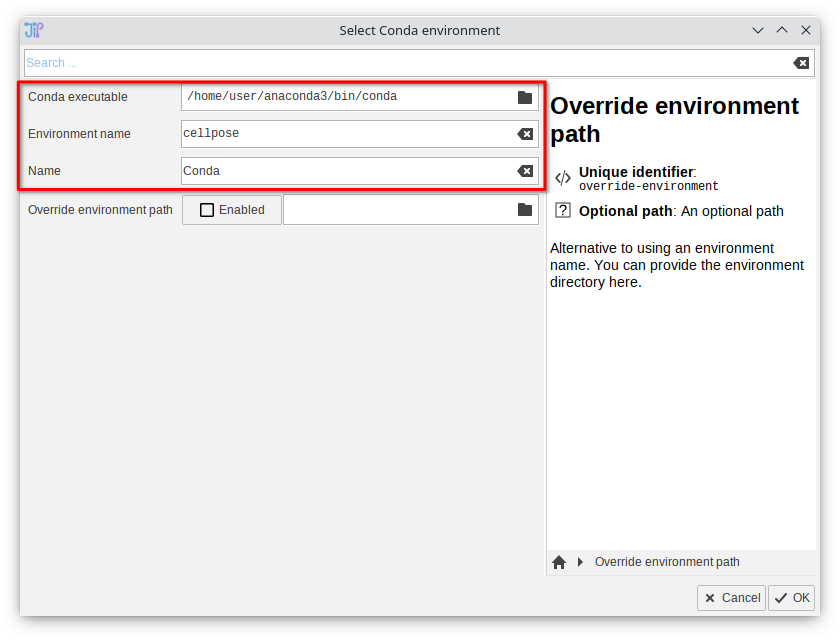
2. Select the Conda executable and environment
Provide the path to the Conda executable and the environment name. Common locations for the executable are:- Windows:
C:/Users/User/anaconda3/Scripts/conda.exe(adapt to your user name) - Linux:
/home/user/anaconda3/bin/conda(adapt to your user name) - MacOS:
/Users/user/opt/anaconda3/bin/conda(adapt to your user name)
cellpose. Adapt the "Environment name" setting accordingly.
Confirm the configuration by clicking "OK".
Installing Cellpose from scratch (automatically)
1. Open the Python integration settings and select the installer
Unless you clicked "Configure Python" in the notification panel, navigate toProject > Application settings > Extensions > Cellpose.
Click the "Select/Install" button in the "Python environment" setting and choose any of the following installation routines:
- Python: Download & install Cellpose (CPU) Will download a portable Python distribution and installs all components of Cellpose via pip. This will install the CPU version of Cellpose.
- Python: Download & install Cellpose (GPU) Will download a portable Python distribution and installs all components of Cellpose via pip. This will install the GPU-accelerated version of Cellpose (please carefully check the pytorch pip package in the configuration dialog that will open)
- Miniconda: Download & install Cellpose (CPU) Will download a Miniconda distribution and installs all components of Cellpose. This will install the CPU version of Cellpose. Currently not actively maintained.
- Miniconda: Download & install Cellpose (GPU) Will download a Miniconda distribution and installs all components of Cellpose. This will install the GPU-accelerated version of Cellpose. Currently not actively maintained.
