Preliminary steps
👉 This tutorial requires that you have installed node template Import images.
If you do not have these templates, you can download them via Manage > Download more templates or by importing the Templates.json file that is provided in the data package. If you do not know how to download or import node templates, please check out our tutorial.
Step 1
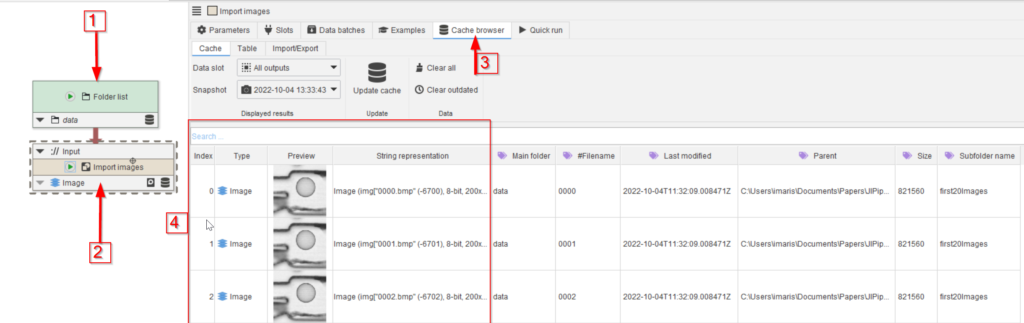
Load a 1-compartment template into JIPipe and add the data folder of the tutorial to the UI (red arrow 1).
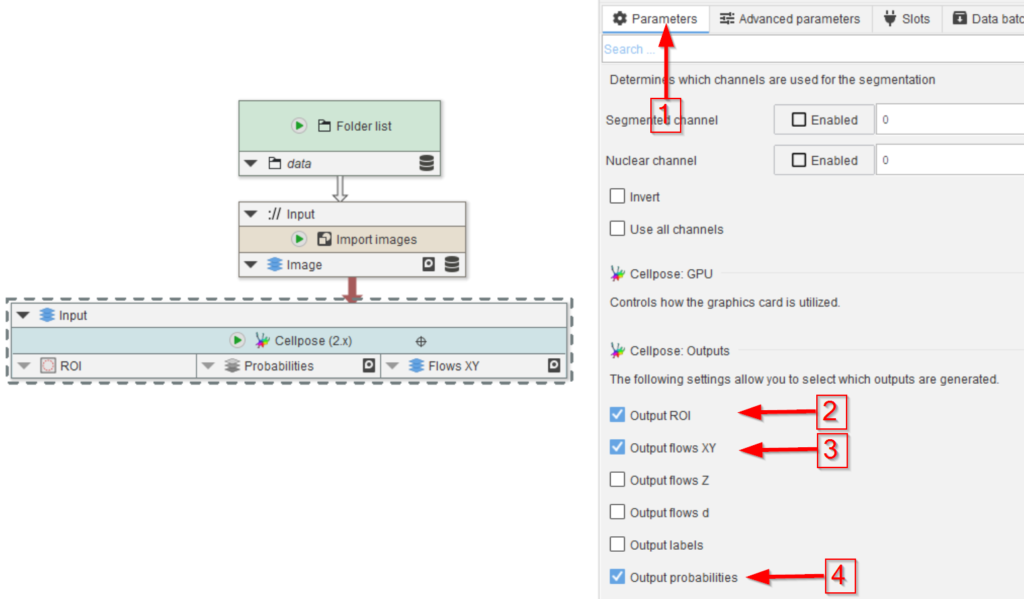
Drag the Import images template node from the Node templates list to the UI and connect it to the Folders node (red arrows 2). Run this node and observe that cache browser (red arrow 3) that now shows 20 images of the droplet example (red rectangle 4).
Step 2
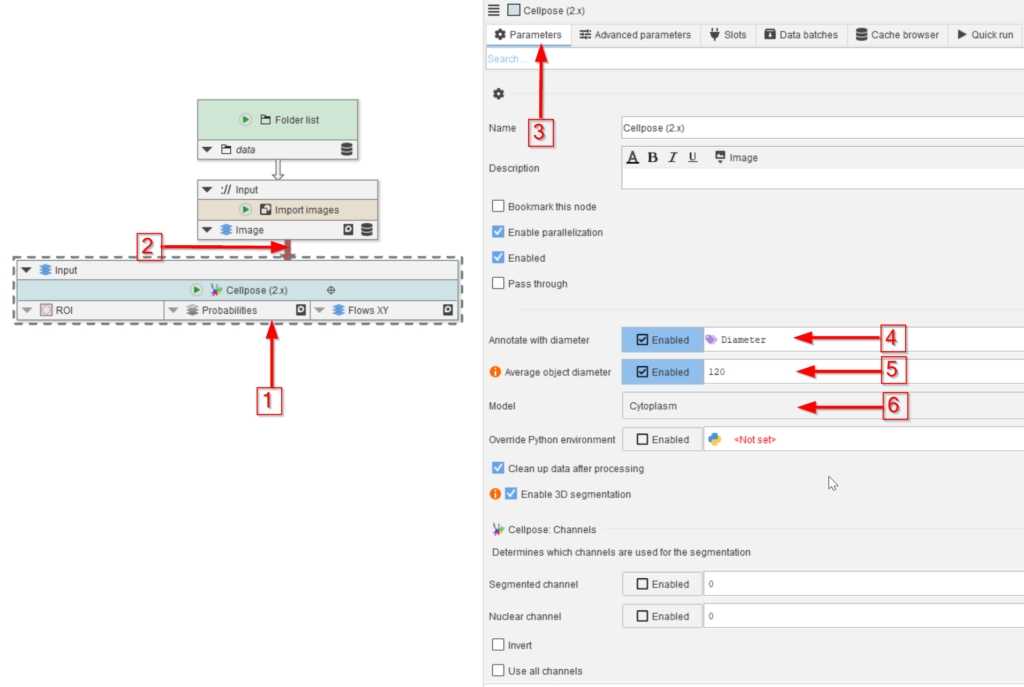
Add a Cellpose (2.x) node to the UI (red arrow 1) and connect it to the Import images node (red arrow 2).
In the Parameters tab (red arrow 3) observe the default annotation Diameter (which is the size guidance for the objects to be found; red arrow 4) and set the Average object diameter parameter to Enabled and its value to 120 (red arrow 5).
Set the segmentation model to Cytoplasm (red arrow 6), which is one of the pre-trained models of Cellpose, and it should work well with the general circular shape of the droplets.
Step 5
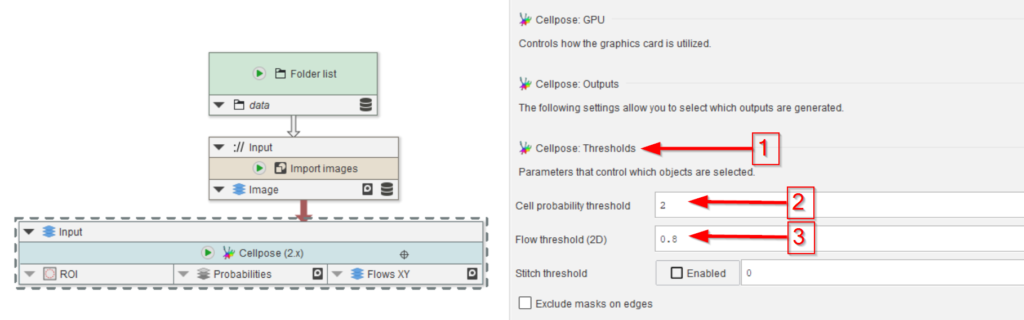
Guidance for these levels can be gained from examining the probability map and identifying the typical levels at the image features of interest.
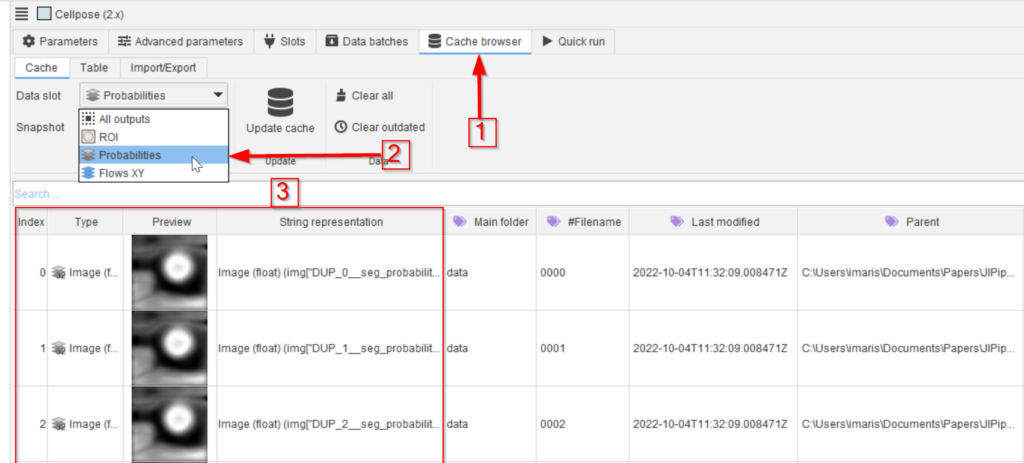
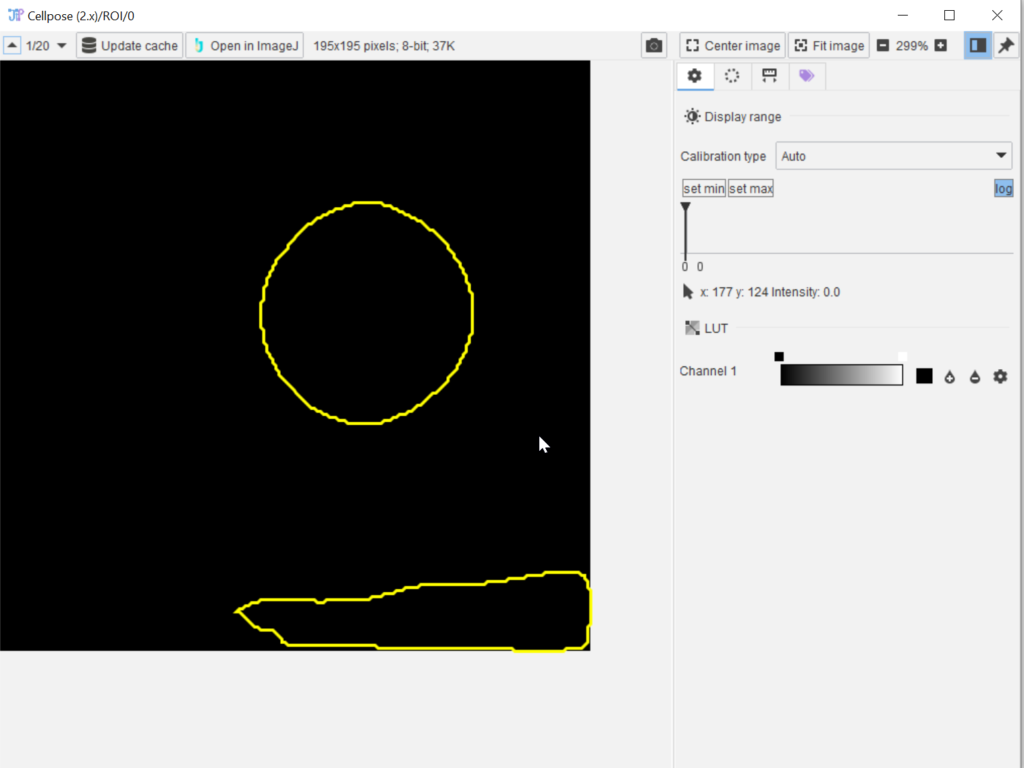
In the Cache browser (red arrow 1) choose the Probabilities data slot (red arrow 2) and examine the outcome (red rectangle 3). Open an item of interest by double-clicking it and use the mouse pointer to observe the values.
Step 10
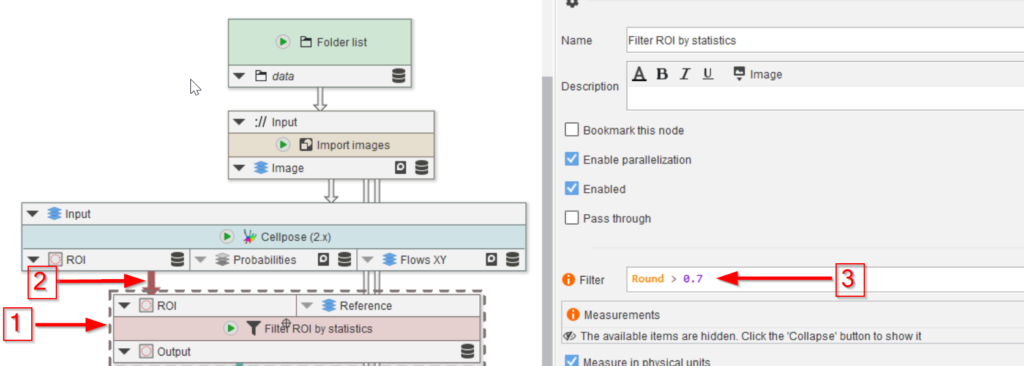
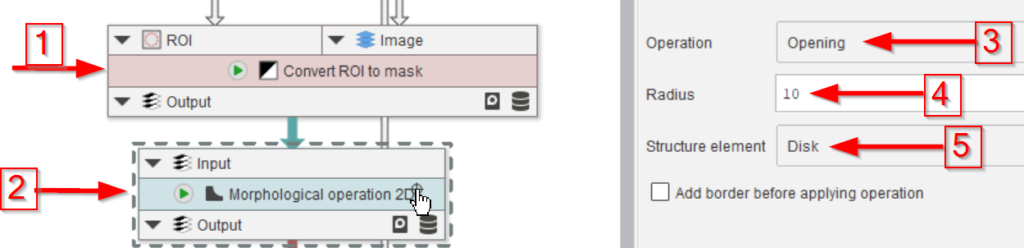
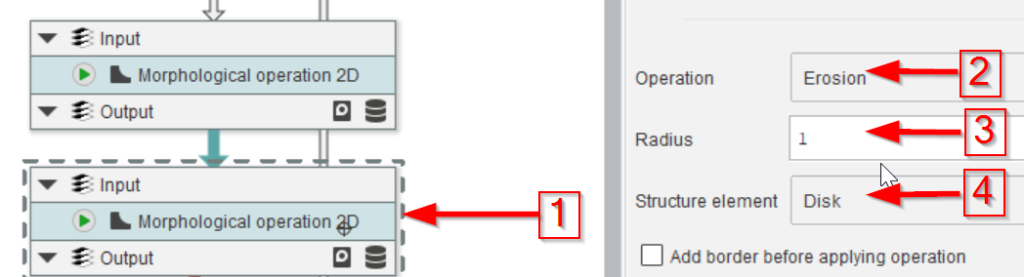
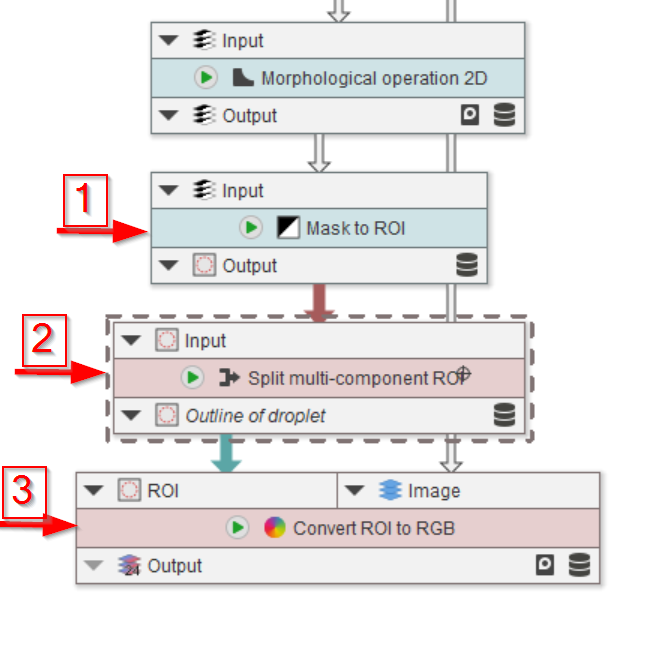
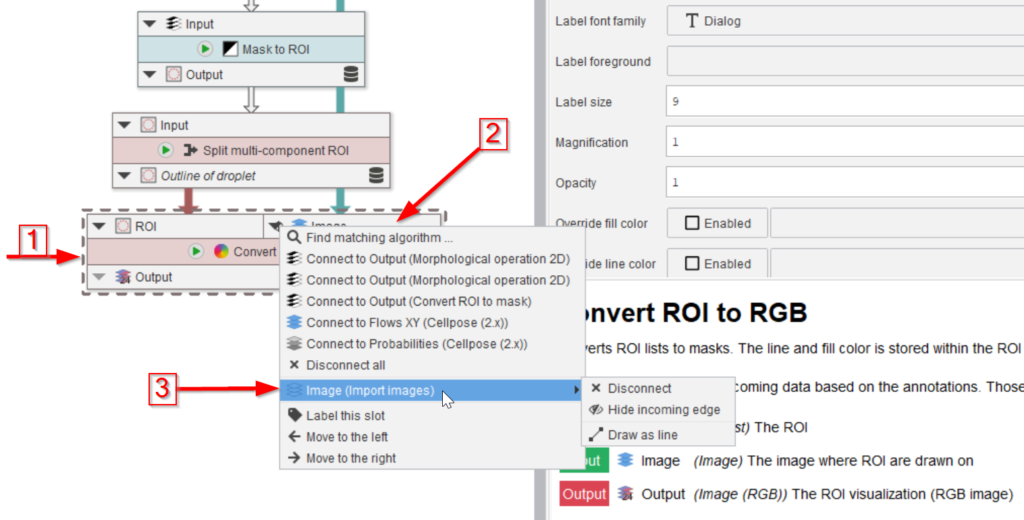
The last three nodes will convert the mask back to ROIs (red arrow 1), split them in case more than one full droplet is found (red arrow 2), and show the quality of the segmentation by overlaying the ROIs with the image (red arrow 3).