R integration
JIPipe provides nodes that allow to execute native R code and use it to process data. All nodes that are relevant to R processing can be found in the category. The following nodes are available:
- R script (iterating) ⭐
(Most commonly used node)
Executes the R script for each iteration step. Each slot receives exactly one data item per step.
- R script (merging)
Executes the R script for each iteration step. Each slot receives multiple data items per step if they cannot be separated by annotations.
Background
The JIPipe R integration basically works by applying the following steps:
Create temporary directories for inputs and outputs
Write all input data to its temporary directory in JIPipe's data table format.
Run a modified R script that has additional variables and functions provided (injected) by JIPipe
Let JIPipe read all data from the output directory. The data must be in JIPipe's data table format.
Writing R scripts
Generally, you can use all R libraries that are available to you in the currently configured R environment. But to facilitate the data transfer between JIPipe and R, you will need to use the functionality provided by the JIPipe-specific R code.
JIPipe-generated variables
On executing the R script, JIPipe will automatically inject the following global variables into the R script:
Name | Description |
|---|---|
| Named list of the number of input data provided by the given input (always 1 for R script (iterating)).
count <- JIPipe.InputSlotRowCounts$Input
|
| Named list of the text annotations associated to the current iteration step.
value <- JIPipe.TextAnnotations$key
|
| Named list of the text annotations associated to the input slot row. This may be helpful if you want to access the annotations before merging them.
value <- JIPipe.InputSlotRowTextAnnotations$key
|
| Named list of variables from the Script variables parameter of the node. |
JIPipe-generated functions
The R script will also have the following functions:
Signature | Description |
|---|---|
| Returns the row storage folder where the data of the given slot and row are stored.
|
| Returns the data of the given input slot and row as data frame. Please note that the input should be a Results table.
|
| Adds a new output folder into the specified output slot and returns the folder path. Optionally, you can assign annotations to add as list of named strings. Please note that the folder must contain data according to the slot's data type (an image file or a CSV file respectively)
|
| Adds the specified data frame into the specified output slot (must be a Results table). Optionally, you can add annotations to the data.
|
| Generates an image file path to be added as output and return the path. Please note that you must use png() or other functions to actually write this file. Optionally, you can add annotations to the data.
|
| Generates an image file path to be added as output and return the path. Please note that you must use tiff() or other functions to actually write this file. Optionally, you can add annotations to the data.
|
Importing data from JIPipe into R
To copy data from the JIPipe into an R variable you will need to use the JIPipe-provided functions to either directly import a data.frame or manually load data from the row storage path.
The JIPipe adapter code provides a convenient function to read a data.frame from a Results table input:
If the input is not a data frame, you can obtain the row folder that contains the file(s) that represent the data.
Exporting data from R into JIPipe
JIPipe expects that data is placed at very specific locations, which can be achieved by utilizing the JIPipe-provided functions and variables.
Depending on which kind of data you want to export, you will have the following options:
You can use a JIPipe-provided function to conveniently output a data.frame back into JIPipe:
For images to be detected by JIPipe, you will need to export an image file (PNG/TIFF/JPG/BMP) into a specific directory. You can obtain the path via the JIPipe-provided functions.
If you are working with other data types, please read about how JIPipe stores this particular type by navigating to and going to the section.
Debugging R scripts
While JIPipe currently does not have any advanced debugging features, you can utilize the log entry generated by JIPipe to identify errors and even run your pipeline outside JIPipe.
- Full script printout
As JIPipe applies modifications to your script, it will print the final script into the log window:
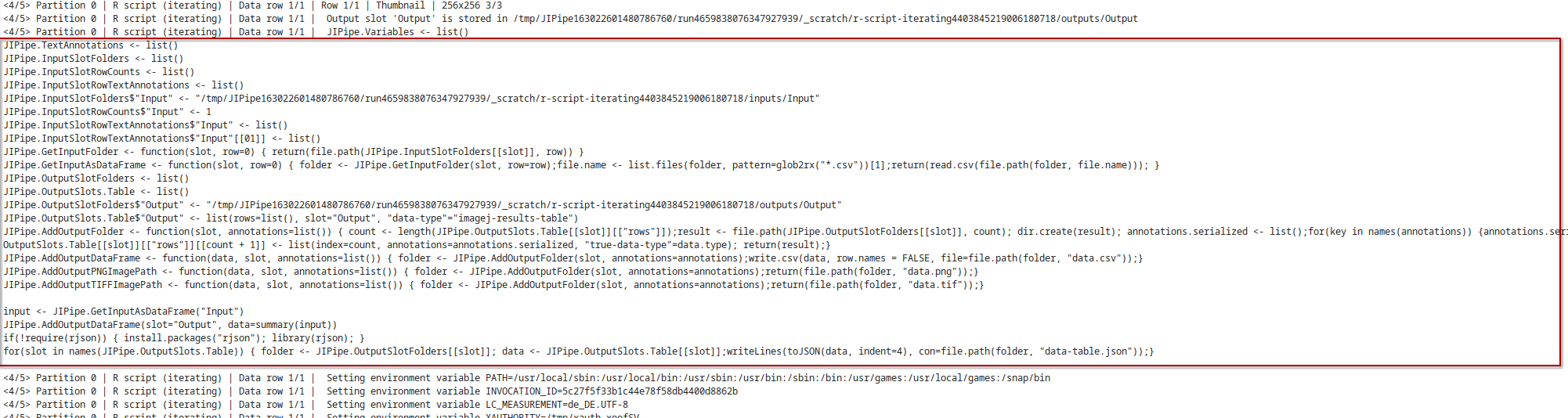
- Commandline run command
JIPipe will also log all environment variables and the full run command:
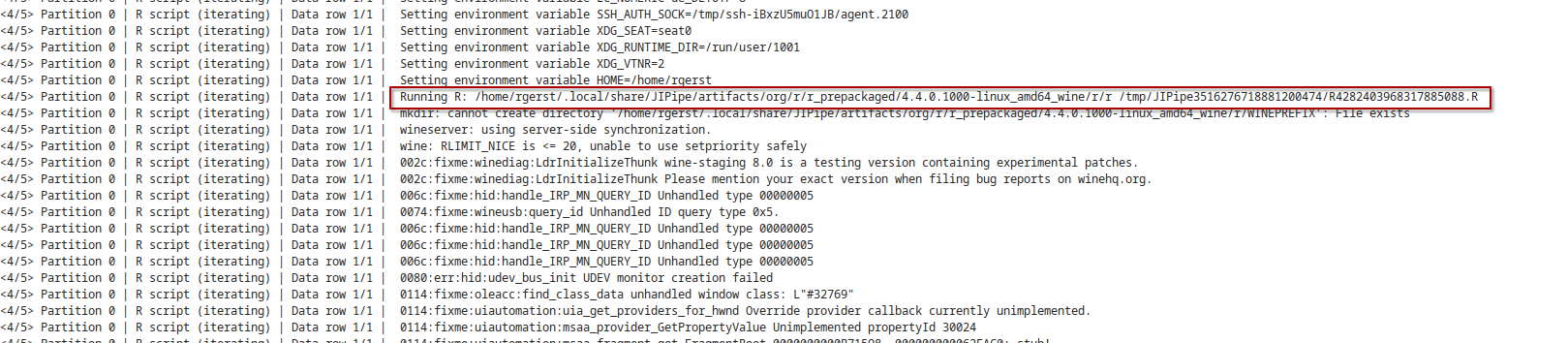
- R stdout/stderr output
JIPipe will also log all standard out and standard error messages directly after the run command.
JIPipe-provided R
JIPipe will automatically take care of setting up R for you and select the newest available artifact for you. The following R versions with the specified libraries are available:
Same packages as 4.4.0.1000.
Overriding the R version
You can always override the utilized R version on a node, project, and application level.
Click the Infos & Settings button at the top right.
Go to the Settings tab on the right-hand side and select .
Ensure that Project default environment is enabled.
Click the Configure ... button and select one of the options.
Select the node you want to change
Go to the panel and activate the parameter .
Click the Configure ... button located next to the parameter and select one of the options.
Navigate to
Select
Click the Configure ... button located next to and select one of the options.